Overview
What is PCoM?
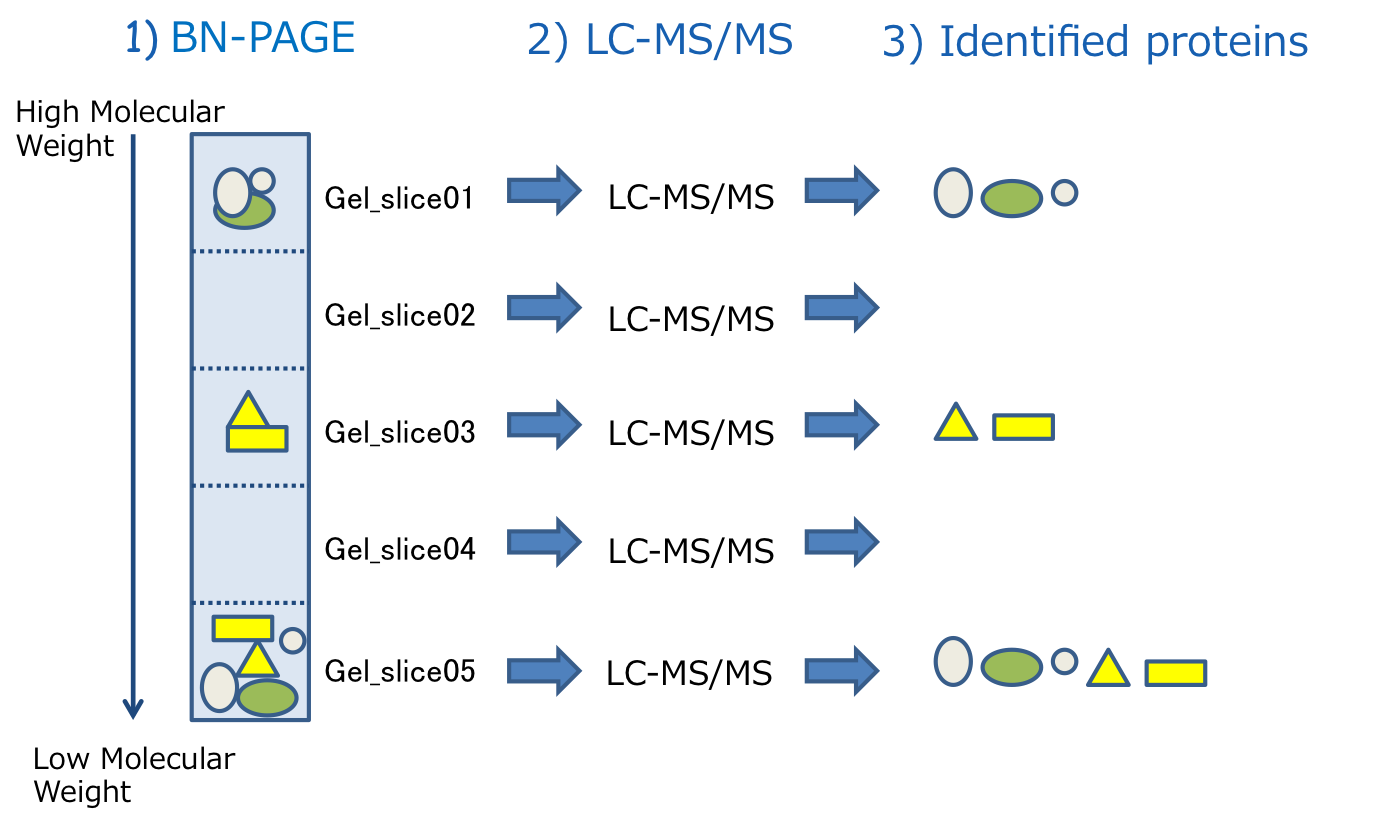
Protein Co-Migration Database for photosynthetic organisms (PCoM) is a powerful tool for predicting the protein complexes in photosynthetic organisms containing your protein of interest. PCoM stores the data obtained by LC-MS/MS analysis of the protein complexes which were separated by BN-PAGE and extracted from BN-PAGE gel slices.
What can users do with PCoM?
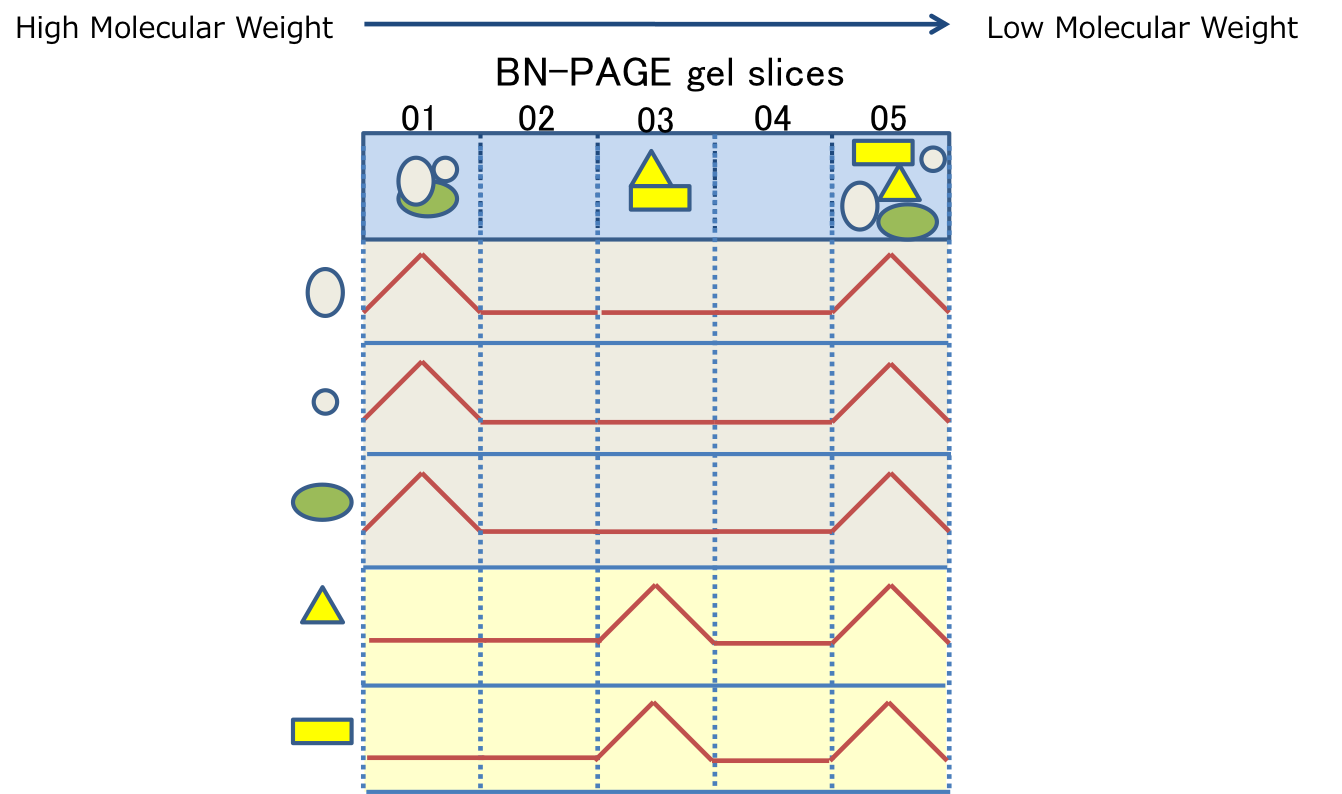
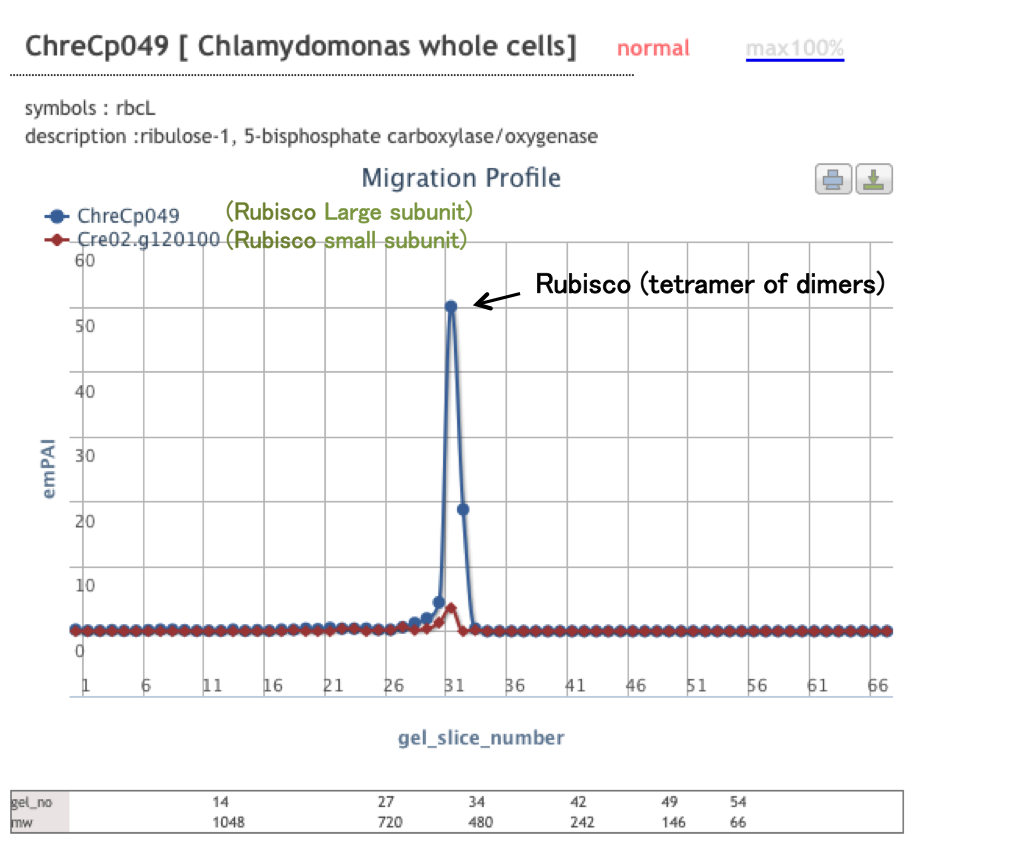
PCoM can show protein migration profiles across the BN-gel slices, which are generated based on the estimation of the protein abundances by the label-free semi-quantitative method, Exponentially modified protein abundance index (emPAI). The migration profile of each protein is good correlated with its band pattern on the BN-PAGE; the gel slice number of each peak is good correlated with the migration distance of the corresponding protein complex and the emPAI of each peak is good correlated with the accumulation level of the corresponding protein complex
Therefore, protein migration profiles provides users following information on protein complexes in model photosynthetic organisms;
1. The number of the protein complexes containing a protein of interest
2. The estimated molecular sizes of the protein complexes containing a protein of interest
3. The relative accumulation levels of each protein complex containing a protein of interest.
In addition, users can obtain the list of the proteins that are possibly interacted with a protein of interest, based on the assumption that the proteins whose peaks in their profiles were overlapped in the same gel slice are possibly interacted each other. Especially, the overall profiles of two proteins are similar, they are possibly the subunits in the same protein complex.
How to use PCoM
Locus ID (for example: AT1G00010 for Arabidopsis, Cre01.g001350 for Chlamydomonas, and sll0611 for Synechocystis PCC6803) can be submitted to PCoM for searching database.
《Search window》 | |
 |
Users can search proteins by typing a Locus ID. Alternatively, users can type a gene symbol (RbcL) or a gene description (Ribulose bisphosphate carboxylase). |
Alternatively, users can select a protein from the protein list including all of the identified proteins by LC-MS/MS. After selecting or searching a protein, the protein migration profile will be displayed. Users can see the list of the proteins identified in each gel slice by clicking on each data point on the profile. After selecting proteins from the list, users can compare the profile of the protein of interest with those of the selected proteins.
References
If you will find that this database is useful, please cite the following publications;
|
|
Atsushi Takabayashi,Ryosuke Kadoya,Masayoshi Kuwano,Katsunori Kurihara,Hisashi Ito,Ryouichi Tanaka and Ayumi Tanaka |
Contact us
![]() Takabayashi@pop.lowtem.hokudai.ac.jp
Takabayashi@pop.lowtem.hokudai.ac.jp